class: center, middle, inverse, title-slide # STATS 250 Lab 05 ## Scatterplots and Linear Regression ### Nick Seewald<br />nseewald@umich.edu ### Week of 09/28/2020 --- class: center, middle # Reminders 💡 Your tasks for the week running Friday 9/25 - Friday 10/2: | Task | Due Date | Submission | |:-----|:---------|:-----------| | Homework 4 | Friday 10/2 8AM ET | course.work | | Lab 5 | Friday 10/2 8AM ET | Canvas | *Stop by office hours! You can attend anyone's -- not just mine!* --- # Lab 3 Comments (Sorry I'm still a bit behind on grading) - Please be careful to answer all parts of every question! - When deciding number of breaks for a histogram, try to avoid empty bins. - Skew direction is which side the tail is on - Skew right implies mean > median; skew left implies mean < median - In Dive Deeper 2, I think we should keep the outlier: there's no reason to believe that William and Mary is fundamentally different from other public schools. - **"Accuracy" or numerical convenience is not a good reason to eliminate a data point.** --- # Homework 3 Summary - **SHOW WORK.** No work = no points 😿 - **Independent events:** `\(P(A \text{ and } B) = P(A)P(B)\)` *if and only if A, B are independent*. Same thing with `\(P(A\mid B) = P(A)\)`. - This must hold *exactly*: 0.786 `\(\neq\)` 0.75 - Events can be mutually exclusive, independent, or neither, but *not both*. - Use numerical support; don't rely on logic. --- class: middle # Weekly Advice - R "draws" graphs like ink on paper. Make a graph (e.g., `plot()`), then use other functions to draw on top of the graph. - Because R draws in "ink", there's no eraser! You need to start over by running `plot()` again. - **The way to get a graphic you like is by trying stuff and adjusting.** - Use R's built-in help for "graphical parameters"! In the console, type `?par`. <img src="https://media1.tenor.com/images/35b9a8d480d756f6d31d2d59d56abb4a/tenor.gif?itemid=5139192" style="width:30%;"></img> --- # Vectors in R (line 59) - A **vector** is a way to hold a collection of things in R. Think of it as a pill organizer. - We can make vectors using the `c()` function. `c` here stands for **c**ombine. ```r x <- c(1, 72.15, -4) x ``` ``` [1] 1.00 72.15 -4.00 ``` --- # `stringsAsFactors` (line 70) ```r penguins <- read.csv("https://raw.githubusercontent.com/STATS250SBI/palmerpenguins/master/inst/extdata/penguins_NArm.csv", * stringsAsFactors = T) ``` - We've got an extra argument to `read.csv()` called `stringsAsFactors`. - Tells `read.csv()` that it should treat data that looks like text as a categorical variable. - In STATS 250, text-like data will almost always be a categorical variable, so we'll be setting `stringsAsFactors = TRUE` often. --- # Scatterplots Revisited (line 82) .pull-left[ ```r plot(bill_depth_mm ~ body_mass_g, data = penguins, main = "Scatterplot of Penguin Body Mass vs. Bill Depth", xlab = "Bill Depth (mm)", ylab = "Body Mass (g)") ``` Notice: 1. "Formula syntax": We specified `y ~ x` in the `plot()` code. 1. Pretty obvious clustering here! **What could be the reason for this?** ] .pull-right[  ] --- # Scatterplots: Color-Coding Points (line 97) .pull-left[ ```r plot(bill_depth_mm ~ body_mass_g, data = penguins, main = "Scatterplot of Penguin Body Mass vs. Bill Depth", xlab = "Bill Depth (mm)", ylab = "Body Mass (g)", * col = c("midnightblue", "brown1", "mediumseagreen")[penguins$species]) ``` - Set `col` argument to a vector of colors - Use `[]` to select color based on categorical variable - Use color **with restraint** ] .pull-right[  ] --- # Color Should Have Meaning .pull-left[ <!-- --> ] .pull-right[ This looks fun, but what does the color *mean*? Color should convey information, and enhance readability.  ] --- # Adding Legends to Plots (line 118) .pull-left[ ```r # Make the plot again plot(bill_depth_mm ~ body_mass_g, data = penguins, main = "Scatterplot of Penguin Body Mass vs. Bill Depth", xlab = "Bill Depth (mm)", ylab = "Body Mass (g)", col = c("midnightblue", "brown1", "mediumseagreen")[penguins$species]) # Add a legend legend("topright", legend = levels(penguins$species), col = c("midnightblue", "brown1", "mediumseagreen"), * pch = 1, title = "Species") ``` ] .pull-right[  ] --- # Plotting Character (`pch`, line 143) .pull-left[ ```r # Make the plot again plot(bill_depth_mm ~ body_mass_g, data = penguins, main = "Scatterplot of Penguin Body Mass vs. Bill Depth", xlab = "Bill Depth (mm)", ylab = "Body Mass (g)", col = c("midnightblue", "brown1", "mediumseagreen")[penguins$species], * pch = c(0, 1, 2)[penguins$species]) # Add a legend legend("topright", legend = levels(penguins$species), col = c("midnightblue", "brown1", "mediumseagreen"), * pch = c(0, 1, 2), title = "Species") ``` ] .pull-right[ 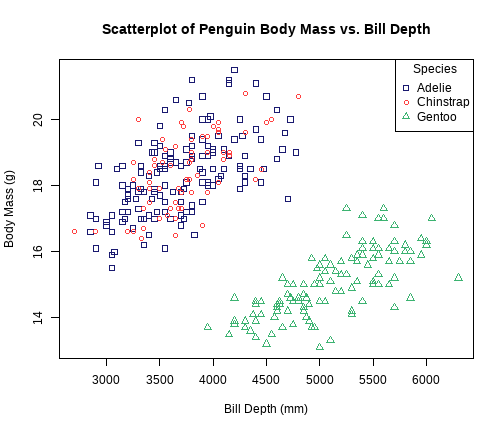 ] --- class: center,middle # Question Break  --- # Correlation (line 165) .pull-left[ Last week's scatterplot: ```r plot(body_mass_g ~ bill_length_mm, data = penguins, main = "Scatterplot of Penguin Body Mass versus Bill Length", xlab = "Bill Length (mm)", ylab = "Body Mass in (g)") ``` ```r cor(penguins$bill_length_mm, penguins$body_mass_g) ``` ``` [1] 0.5894511 ``` ] .pull-right[ 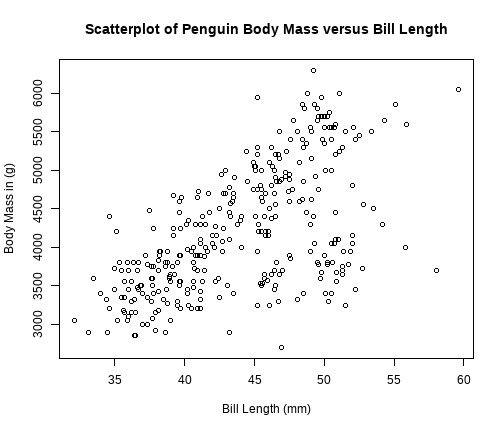 ] --- # Correlation Matrices (line 183) First, subset the data to just look at quantitative variables, then feed that subset to `cor()` to compute a *correlation matrix* ```r numericPenguins <- subset(penguins, select = c("bill_length_mm", "bill_depth_mm", "flipper_length_mm", "body_mass_g")) cor(numericPenguins) ``` ``` bill_length_mm bill_depth_mm flipper_length_mm body_mass_g bill_length_mm 1.0000000 -0.2286256 0.6530956 0.5894511 bill_depth_mm -0.2286256 1.0000000 -0.5777917 -0.4720157 flipper_length_mm 0.6530956 -0.5777917 1.0000000 0.8729789 body_mass_g 0.5894511 -0.4720157 0.8729789 1.0000000 ``` Each "entry" in the correlation matrix is the correlation between the variables labeling that entry's row and column. --- # Linear Regression (line 197) <!-- To run linear regression use `lm()` and formula syntax (`y ~ x`) --> ```r reg1 <- lm(body_mass_g ~ bill_length_mm, data = penguins) summary(reg1) ``` ``` Call: lm(formula = body_mass_g ~ bill_length_mm, data = penguins) Residuals: Min 1Q Median 3Q Max -1759.38 -468.82 27.79 464.20 1641.00 Coefficients: Estimate Std. Error t value Pr(>|t|) (Intercept) 388.845 289.817 1.342 0.181 bill_length_mm 86.792 6.538 13.276 <2e-16 *** --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 Residual standard error: 651.4 on 331 degrees of freedom Multiple R-squared: 0.3475, Adjusted R-squared: 0.3455 F-statistic: 176.2 on 1 and 331 DF, p-value: < 2.2e-16 ``` --- # ANOVA Tables (line 214) Give your regression model (ours is `reg1`) to the `anova()` function: ```r anova(reg1) ``` ``` Analysis of Variance Table Response: body_mass_g Df Sum Sq Mean Sq F value Pr(>F) bill_length_mm 1 74792533 74792533 176.24 < 2.2e-16 *** Residuals 331 140467133 424372 --- Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1 ``` `$$R^2 = \frac{\text{SSM}}{\text{SST}}$$` --- class:inverse # Lab Project ⌨️ You will be **randomly** moved to a breakout room for the rest of the lab (minus ~10 minutes) .pull-left[ ### Your tasks 1. Introduce yourself to your collaborators! 1. **Work together** to complete the "Try It!" and "Dive Deeper" portions of the lab assignment by copy/pasting and modifying appropriate code from earlier in the document. ] .pull-right[ ### How to get help - I'll be floating around between breakout rooms to check on everyone - Use the "Ask for help" button to flag me down - Let me know when you're done ] --- class: center, middle, inverse # What questions do you have? Any issues? --- class: center, middle # "Exit Ticket" Please take 1-2 minutes to complete the survey at ### [bit.ly/250ticket5](http://bit.ly/250ticket5) --- # Reminders 💡 Your tasks for the week running Friday 9/25 - Friday 10/2: | Task | Due Date | Submission | |:-----|:---------|:-----------| | Homework 4 | Friday 10/2 8AM ET | course.work | | Lab 5 | Friday 10/2 8AM ET | Canvas | *Stop by office hours! You can attend anyone's -- not just mine!*